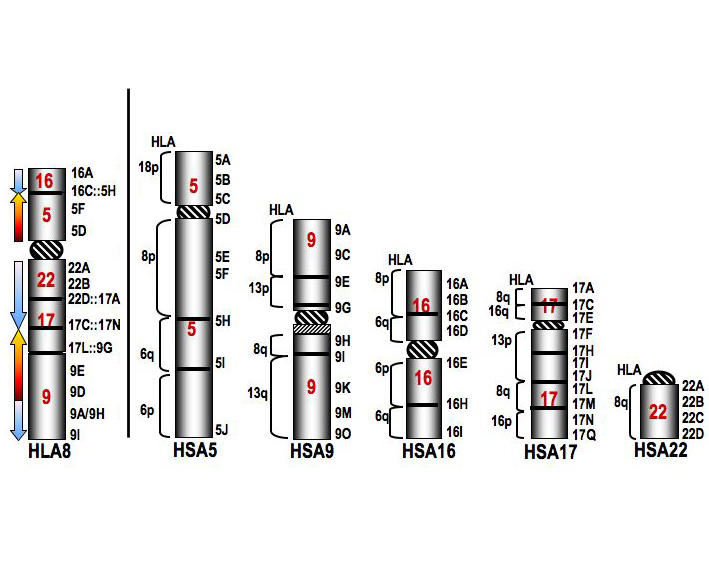
 |
|
|
CODE |
PROBE
|
ACC.N
|
MAP
|
UCSC May 2004
|
REMARKS
|
|
|
16A
|
RP11-614O5 |
BES |
16p13.13 |
chr16:10,455,533-10,648,602 |
|
|
16B
|
RP11-614O5 |
AQ361328 |
16p13.13 |
chr16:10,455,533-10,648,602 |
|
|
|
|
RP11-449C8 |
BES |
16p13.12 |
chr16:14,275,081-14,463,453 |
|
|
|
|
RP11-44P2 |
BES |
16p12.13 |
chr16:19,163,212-19,355,377 |
|
|
16C
|
RP11-467J24 |
|
16p12.3 |
chr16:19,290,201-19,451,651 |
HLA8p&6qcen |
| B-A |
NLE
|
CH271-279L5 |
BES |
|
chr5:75,621,759/chr16:19,285,157 |
HyA translocation HLA8p |
| CH271-276H20 |
BES |
|
chr5:75,638,548/chr16:19,300,749 |
|
5H
|
RP11-204O23 |
BES |
|
chr5:75,668,456-75,845,041 |
HLA8p&6qcen |
|
|
|
RP11-944F8 |
BES |
5q13.3 |
chr5:75,479,146-75,660,029 |
|
|
|
|
RP11-172K14 |
AFMA348WD1 |
5q13.3 |
chr5:74,563,429-74,563,728 |
|
|
|
5G
|
RP11-480H11 |
AC016643 |
5q12.3 |
chr5:65,059,160-65,239,118 |
|
|
|
5F
|
RP11-105M3 |
BES |
5q11.2 |
chr5:54,132,527-54,318,331 |
|
|
5E |
RP11-160F8 |
AC034246 |
5q11.2 |
chr5:53,365,247-53,520,260 |
|
|
5D |
RP11-317O24 |
BES |
|
chr5:50,100,508-50,293,531 |
|
|
|
|
CENTROMERE
|
|
|
|
|
| B-L |
|
|
|
|
|
HLA fusion |
|
|
22A
|
CTA-115F6 |
BES |
22q11.2 |
chr22:16,153,880-16,340,333 |
|
|
|
|
CTA-322B1 |
BES |
22q11.23 |
chr22:22,639,817-22,716,609 |
|
 |
22B |
CTA-57G9 |
BES |
22q12.1 |
chr22:27,837,088-27,950,960 |
|
| B-N |
NLE |
CH271-340F4 |
BES |
|
chr22:31,155,960/chr4:140.754.577 |
HLA8q&2q |
|
|
22C
|
CTA-415G2 |
BES |
22q12.3 |
chr22:31,690,491-31,820,448 |
|
|
|
|
CTA-250D10 |
BES |
22q13.2 |
chr22:40,577,264-40,798,159 |
|
|
|
22D
|
RP5-925J7 |
AL078622 |
22q13.32 |
chr22:47,675,206-47,773,652 |
|
| B-L |
|
|
|
|
|
HLA fusion |
|
|
17A
|
RP11-411G7 |
AC027455 |
17p13.3 |
chr17:427,025-572,435 |
|
|
|
|
RP11-655D3 |
AC026591 |
17p13.1 |
chr17:9,660,550-9,814,021 |
|
|
|
17B
|
RP11-746E8 |
BES |
17p12 |
chr17:12,435,731-12,620,614 |
|
|
|
|
RP11-803H14 |
BES |
17p11.2 |
chr17:16,396,847-16,555,073 |
|
 |
17C |
RP11-92B11 |
|
17p11.2 |
chr17:16,467,786-16,646,713 |
HLA8q&16p |
|
B-A
|
|
|
|
|
|
HyA inversion
|
|
|
17N
|
RP11-346K10 |
BES |
17q24.1 |
chr17:61,533,516-61,702,132 |
HLA8q&16p |
| B-N |
NLE |
CH271-32K04 |
BES |
|
chr17:59,288,449/chr2:27,705,953 |
HLA8q&10p |
 |
17M |
RP11-630H24 |
|
17q23.3 |
chr17:59,198,386-59,358,882 |
|
|
|
|
RP11-1122L1 |
BES |
17q23.3 |
chr17:58,102,921-58,244,753 |
|
|
|
17L
|
RP11-464D20 |
AC008026 |
17q23.3 |
chr17:57,765,672-57,984,805 |
|
|
B-A
|
|
|
|
|
|
HyA translocation
|
|
9G |
RP11-671K21 |
BES |
9p13.2 |
chr9:38,241,473-38,423,166 |
HLA8q&13q |
| B-A |
NLE |
CH271-169O22 |
BES |
|
chr9:38,763,347/chr9:27,065,324 |
HyA inversion HLA8q |
| CH271-163A2 |
BES |
|
chr9:38,746,375/chr9:27,098,102 |
|
|
9E
|
RP11-1006E22 |
BES |
9p21.2 |
chr9:27,142,243-27,331,367 |
HLA8q&13p |
|
|
|
RP11-784B21 |
BES |
9p21.3 |
chr9:24,745,524-24,913,097 |
|
|
|
9D
|
RP11-296K11 |
BES |
9p21.3 |
chr9:22,245,370-22,411,015 |
|
| B-N |
NLE |
CH271-131G15 |
BES |
|
chr9:22,241,811/chr9:111,392,119 |
HLA8q&13q |
|
|
|
RP11-101L23 |
BES |
9p22.1 |
chr9:19,132,752-19,329,389 |
|
|
|
9C
|
RP11-58K1 |
BES |
9p22.3 |
chr9:15,874,140-16,051,195 |
|
 |
9B |
RP11-472F14 |
AL353718 |
9p24.1 |
chr9:6,454,979-6,601,726 |
|
 |
|
RP11-443B9 |
AL591644 |
9p24.3 |
chr9:1,901,824-1,999,570 |
|
 |
9A |
RP11-143M15 |
BES |
9p24.3 |
chr9:812,146-991,152 |
|
| B-H |
|
|
|
|
|
HSA inversion |
 |
9H |
RP11-203L2 |
BES |
9q21.1 |
chr9:68,528,294-68,682,365 |
|
 |
|
RP11-69B23 |
BES |
9q21.1 |
chr9:68,609,860-68,767,622 |
|
 |
9I
|
RP11-876N18 |
|
9q13 |
chr9:68,871,474-69,076,493 |
HLA8q&13q |
|
B-A
|
|
|
|
|
|
HyA inversion
|
|

