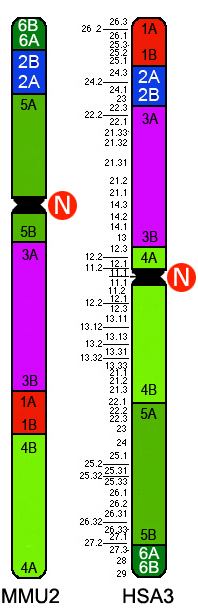
|
|
BAC | Accession # | map | UCSC March 2006 |
comments
|
||
 |
6B
|
RP11-313F11 | AC016953 | 3q29 | chr3:196,907,957-197,370,697 | ||
 |
6
|
RP11-201D16 | AC079623 | 3q28 | chr3:193,773,254-193,974,704 | ||
 |
6
|
RP11-298A18 | AC063932 | 3q28 | chr3:189,731,862-189,918,758 | ||
 |
6
|
RP11-88P6 | AC018919 | 3q27.3 | chr3:188,693,472-188,860,113 | ||
 |
6
|
RP11-42D20 | AC007690 | 3q27.3 | chr3:188,091,027-188,272,557 | ||
 |
6A
|
RP11-177B11 | BES | 3q27.3 | chr3:187,819,875-187,998,697 |
splits
|
|
 |
2B
|
RP11-240N7 | AC098648 | 3p22.3 | chr3:36,506,239-36,658,135 |
splits
|
|
 |
2
|
RP11-607P24 | BES | 3p22.3 | chr3:36,298,506-36,506,070 | ||
 |
2
|
RP11-437B5 | AC105901 | 3p22.3 | chr3:36,110,125-36,296,348 | ||
 |
2
|
RP11-802J21 | AC109585 | 3p22.3 | chr3:35,107,296-35,320,376 | ||
 |
2
|
RP11-952G4 | AC123901 | 3p23 | chr3:33,814,936-34,032,343 | ||
 |
2
|
RP11-627J17 | AC112211 | 3p23 | chr3:32,980,609-33,163,117 | ||
 |
2
|
RP11-722P17 | AC097361 | 3p24.1 | chr3:28,937,458-29,112,483 | ||
 |
2
|
RP11-109D5 | AC074287 | 3p24.2 | chr3:25,497,571-25,696,055 | ||
 |
2
|
RP11-263O21 | AC097635 | 3p25.1 | chr3:19,874,965-20,039,419 | ||
 |
2
|
RP11-421B21 | AC090949 | 3p25.1 | chr3:15,147,209-15,324,528 | ||
 |
2A
|
RP11-616M11 | AC090954 |
3p24
|
chr3:15,045,785-15,213,797
|
splits
|
|
 |
5A
|
at 6.93 Kb from 4B
|
|||||
 |
5
|
RP11-787P10 | BES | 3q22.1 | chr3:131,347,364-131,500,312 | ||
 |
5
|
RP11-893G19 | BES | 3q22.1 | chr3:131,462,583-131,646,610 | ||
 |
5
|
RP11-21N8 | AC023438 | 3q22.1 | chr3:131,810,596-131,961,213 | ||
 |
5
|
RP11-505J9 | AC024897 | 3q24 | chr3:149,845,113-150,049,906 | ||
 |
5
|
RP11-355I21 | AC025826 | 3q26.1 | chr3:163,822,353-164,122,697 | ||
 |
MMU cen (neocentromere) | ||||||
 |
5
|
RP11-418B12 | AC079910 | 3q26.1 | chr3:164,539,723-164,707,155 | ||
 |
5
|
RP11-526M23 | AC048332 | 3q26.1 | chr3:166,898,263-167,089,798 | ||
 |
5
|
RP11-114M1 | AC026355 | 3q26.32 | chr3:178,755,557-178,912,996 | ||
 |
5
|
RP11-436A20 | AC008009 | 3q26.32 | chr3:182,104,591-182,316,939 | ||
 |
5
|
RP11-329B9 | AC107294 | 3q26.33 | chr3:185,897,030-186,090,450 | ||
 |
5
|
RP11-218A22 | AC108670 | 3q27.2 | chr3:186,915,322-187,078,478 | ||
 |
5
|
RP11-443P15 | AC108671 | 3q27.2 | chr3:187,095,608-187,269,476 | ||
 |
5
|
RP11-1118G15 | BES | 3q27.2 | chr3:187,280,861-187,431,522 | ||
 |
5
|
RP11-379C23 | AC007917 | 3q27.2 | chr3:187,368,240-187,555,207 | ||
 |
5
|
RP11-709J22 | BES | 3q27.3 | chr3:187,553,921-187,754,882 | ||
 |
5B
|
RP11-177B11 | BES | 3q27.3 | chr3:187,819,875-187,998,697 |
splits
|
|
| CH250-453O21 | splits on HSA at ~36/~187Mb | ||||||
 |
3A
|
RP11-240N7 | AC098648 | 3p22.3 | chr3:36,506,239-36,658,135 |
splits
|
|
 |
3
|
RP11-640L9 | AC105749 | 3p22.3 | chr3:36,652,404-36,852,853 | ||
 |
3
|
RP11-129K12 | AC011816 | 3p22.3 | chr3:36,866,838-37,036,150 | ||
 |
3
|
RP11-491D6 | AC006583 | 3p22.3 | chr3:37,034,150-37,136,520 | ||
 |
3
|
RP11-395P16 | AC130472 | 3p21.31 | chr3:47,584,747-47,778,906 | ||
 |
3
|
RP11-380J21 | AC092419 | 3p14.1 | chr3:64,053,486-64,212,751 | ||
 |
3
|
RP11-634L22 | BES | 3p12.3 | chr3:75,452,260-75,628,601 | ||
 |
3B
|
at 70 Kb from 4A
|
|||||
 |
1A
|
at 4.328 Kb from the human 3p telomere
|
|||||
 |
1
|
RP11-183N22 | AL512885 | 3p26.1 | chr3:4,328,222-4,493,696 | ||
 |
1
|
RP11-542K24 | AC066598 | 3p26.1-25.3 | chr3:8,060,757-8,261,100 | ||
 |
1
|
RP11-732C9 | BES | 3p25.2 | chr3:12,421,029-12,628,309 | ||
 |
1
|
RP11-588P9 | AC018836 | 3p25.2 | chr3:12,791,778-12,976,492 | ||
 |
1
|
RP11-316A10 | AC090937 | 3p25.1 | chr3:14,886,277-15,046,975 | ||
 |
1B
|
RP11-616M11 | AC090954 | 3p24 | chr3:15,045,785-15,213,797 |
splits
|
|
| CH250-398B15 | splits on HSA at ~15/~131Mb | ||||||
 |
4B
|
at 6.93 Kb from 5A
|
|||||
 |
4
|
RP11-924M2 | BES | 3q22.1 | chr3:131,126,901-131,354,303 |
dup
|
|
 |
4
|
RP11-124N2 | AC079848 | 3q21.2 | chr3:127,272,174-127,434,156 | ||
 |
4
|
RP11-757I12 | AC092908 | 3q21.1 | chr3:123,727,494-123,901,752 | ||
 |
4
|
RP11-305I9 | AC092981 | 3q13.33 | chr3:120,465,181-120,625,176 | ||
 |
4
|
RP11-454H13 | AC084198 | 3q12.3 | chr3:102,798,688-102,993,642 | ||
 |
4
|
HSA cen (neocentromere) | |||||
 |
4
|
RP11-655A17 | AC107204 | 3p11.1 | chr3:87,099,705-87,270,490 | ||
 |
4
|
RP11-536K4 | AC016942 | 3p12.3 | chr3:76,682,896-76,834,909 | ||
 |
4
|
RP11-601J5 | BES | 3p12.3 | chr3:75,877,002-76,036,449 | ||
 |
4
|
RP11-413E6 | AC108724 | 3p12.3 | chr3:75,698,634-75,885,406 |
MS (1)
|
|
 |
4
|
RP11-1053M22 | BES | 3p12.3 | chr3:75,588,180-75,802,856 |
NS (2)
|
|
 |
4A
|
at 70 Kb from 3B
|
|||||
COMMENTS
(1) RP11-413E6. The 4A region became telomeric following the fission that generated an autonomous chromosome 21 in Catarrhini ancestor.
It became internal in HSA-PTR-GGO ancestor following a pericentric inversion. The signals on multiple telomeric sites is, therefore, expected.
(2) RP11-1053M22 failed to yield any FISH signal. This is a special region, full of segmental duplications that, probably, are human (or great apes) specific.