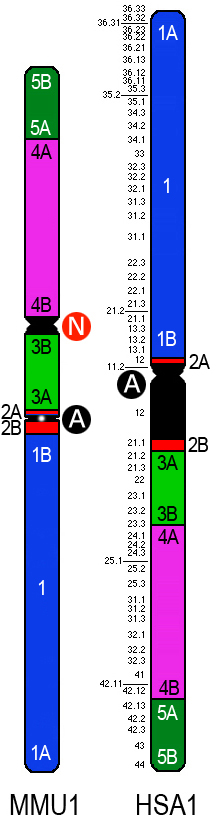
 |
|
|
|
BAC |
Accession # |
map |
UCSC March 2006
|
comments |
 |
5B
|
RP11-438F14 |
AC098483 |
1q44 |
chr1:246,754,133-246,932,000 |
|
 |
5
|
RP11-385F5 |
AL359921 |
1q43 |
chr1:234,752,824-234,966,581 |
|
 |
5
|
RP11-3K22 |
AL359874 |
1q42.13 |
chr1:227,082,837-227,162,782 |
|
 |
5A
|
RP4-621O15 |
AL713899 |
1q42.13 |
chr1:226,810,735-226,866,653 |
splits |
|
|
CH250-471F6 |
|
|
|
signals on HSA at ~161/~226Mb; dup on MMU |
|
|
4A
|
RP11-331H2 |
AL392003 |
1q23.3 |
chr1:161,033,866-161,225,664 |
overlaps 3B |
 |
4
|
RP11-332H17 |
AL356475 |
1q24.2 |
chr1:168,242,366-168,350,412 |
|
 |
4
|
RP11-152A16 |
BES |
1q25.2 |
chr1:177,339,824-177,521,905 |
|
 |
4
|
RP11-134C1 |
BES |
1q31.1 |
chr1:186,252,053-186,417,889 |
|
 |
4
|
RP11-553K8 |
AL157402 |
1q31.3 |
chr1:196,750,597-196,960,927 |
|
|
|
4
|
RP11-57I17 |
AL137789 |
1q32.2 |
chr1:205,811,787-205,957,118 |
|
 |
4
|
RP11-123O6 |
BES |
1q32.2 |
chr1:209,402,692-209,548,350 |
|
 |
4
|
RP11-74E6 |
BES |
1q41 |
chr1:212,560,677-212,729,794 |
|
 |
4
|
RP11-324K19 |
AC118468 |
1q41 |
chr1:219,203,440-219,218,989 |
|
 |
4B
|
RP4-621O15 |
AL713899 |
1q42.13 |
chr1:226,810,735-226,866,653 |
splits |
|
|
|
MMU cen (neocentromere) |
|
 |
3B
|
RP11-572K18 |
AL445197 |
1q23.3 |
chr1:160,918,751-161,035,790 |
overlaps 4A |
 |
3
|
RP11-8D14 |
AC068728 |
1q23.3 |
chr1:158,924,552-159,088,467 |
|
 |
3
|
RP11-98F1 |
AL353760 |
1q22 |
chr1:153,539,660-153,543,654 |
|
 |
3
|
RP11-458I7 |
fishClone |
1q21.2 |
chr1:148,238,619-148,426,829 |
|
 |
3
|
RP11-353N4 |
fishClone |
1q21.1-2 |
chr1:147,853,299-147,978,467 |
dup on HSA |
 |
3A |
RP11-14N7 |
AL513526 |
1q21.1 |
chr1:147,156,861-147,214,458 |
dup on HSA |
 |
2A |
RP11-453B6 |
BX248407 |
1p11.1–11.2 |
chr1:121,045,874-121,186,957 |
dup on HSA; no signal on MMU |
 |
2
|
HSA cen (inactivated, ancestral to MMU)*
|
|
 |
2
|
RP11-35B4 |
AL359093 |
1q21.1+1p11+1p36 |
chr1:144,000,592-144,166,607 |
|
 |
2 |
RP11-30I17 |
BES |
1q21.1 |
chr1:144,174,052-144,361,891 |
|
 |
2 |
RP11-314N2 |
AL445591 |
1q21.1 |
chr1:145,732,430-145,872,521 |
dup on HSA; dup on MMU |
 |
2B |
RP11-763B22 |
AL592492 |
1q21.1 |
chr1:147,000,772-147,158,860 |
dup on HSA; dup on MMU |
 |
1B |
RP11-439A17 |
fishClone |
1p11.2 |
chr1:120,548,680-120,738,218 |
|
 |
1
|
RP5-1042I8 |
AL359752 |
1p11.2-12 |
chr1:120,134,618-120,272,572 |
|
 |
1
|
RP11-192J8 |
BES |
1p12 |
chr1:118,169,383-118,169,829 |
|
 |
1
|
RP11-284N8 |
AL365361 |
1p13.3 |
chr1:110,897,276-111,090,458 |
|
 |
1
|
RP11-138K16 |
AC093559 |
1p21.2 |
chr1:99,728,340-99,904,318 |
|
 |
1
|
RP11-324C23 |
AL445992 |
1p22.1 |
chr1:91,951,846-91,986,170 |
|
 |
1
|
RP11-302M6 |
AL391497 |
1p22.2 |
chr1:90,062,248-90,252,009 |
|
 |
1
|
RP11-254E16 |
BES |
1p31.1 |
chr1:84,539,796-84,689,450 |
|
 |
1
|
RP11-316C12 |
AL627317 |
1p31.1 |
chr1:71,621,580-71,715,216 |
|
 |
1
|
RP11-195G13 |
BES |
1p32.3 |
chr1:54,100,318-54,292,540 |
|
 |
1
|
RP5-1154B21 |
(D1S3315) |
1p33 |
chr1:47,605,724-47,806,009 |
|
 |
1
|
RP11-266K22 |
AL451070 |
1p35.2 |
chr1:31,512,242-31,645,345 |
|
 |
1
|
RP11-265F14 |
AL512883 |
1p32.21 |
chr1:15,630,693-15,735,006 |
|
 |
1A
|
RP11-421C4 |
BES |
1p36.33 |
chr1:1,247,484-1,432,829 |
|
* A human-specific pericentric inversion is present on this chromosome, but it involves
only pericentromeric duplications (Szamalek et al. Hum. Genet. 120:126-138, 2006)
|

