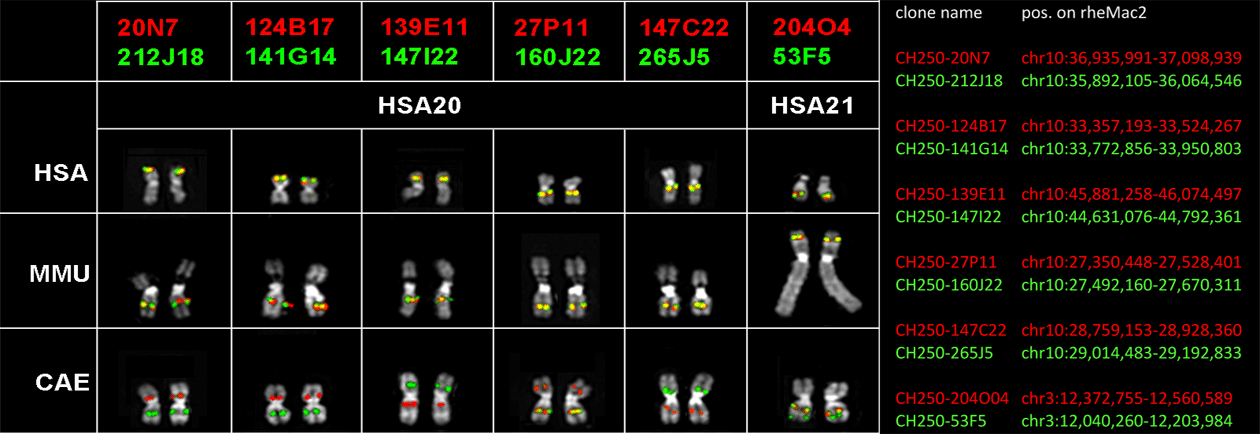
|
|
sequence orientation with respect to HSA |
|
|
centromere |
|
|
neocentromere |
|
|
sinteny breaks occurred in the vervet lineage |
|
|
sinteny breaks occurred in the human lineage |
|
|
duplicated BAC clones |
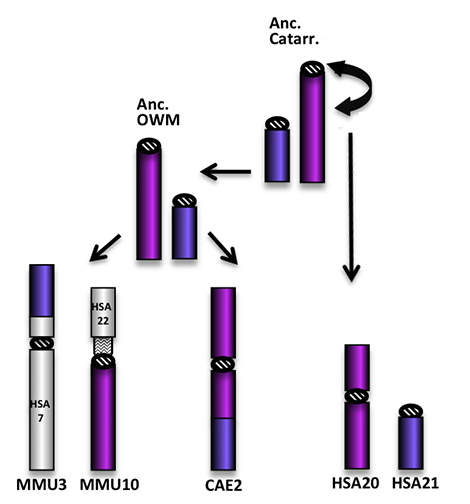
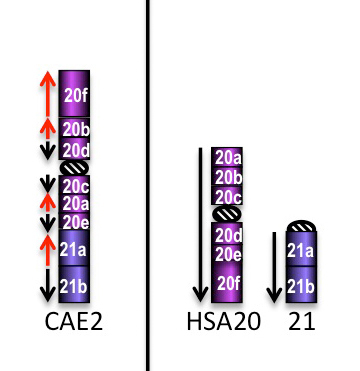
CAE2 is the result of complex successive inversions, not reported in the Figure above
|
|
|
HSA chr. |
clone |
Acc.N. |
UCSC Feb.2009 (hg19)
|
map |
note
|
|
|
20f |
RP11-476I15 |
AL137028.9 |
chr20:62,906,096-62,965,520 |
20q13.33 |
|
|
|
20f |
RP5-1059L7 |
AL121913 |
chr20:56,232,155-56,382,378 |
|
|
|
|
20f |
RP5-906C1 |
AL133342 |
chr20:47,395,324-47,506,137r |
|
|
|
|
20f |
RP11-257H6 |
BES |
chr20:40,066,498-40,244,105 |
|
|
|
|
20f |
CH250-27P11 |
|
chr20:35,554,047-35,741,568 |
|
macaque clone
|
|
|
20f |
RP11-1152L20 |
BES |
chr20:35,651,140-35,776,134 |
20q11.23 |
|
|
|
-
|
|
|
|
|
|
|
|
20b |
RP11-643H5 |
BES |
chr20:11,431,493-11,596,313 |
20p12.2 |
|
|
|
20b |
CH250-147I22 |
|
chr20:11,323,402-11,476,445 |
|
macaque clone
|
|
|
20b |
RP11-975D16 |
BES |
chr20:11,287,806-11,478,681 |
|
|
|
|
20b |
RP11-43J1 |
BES |
chr20:8,598,111-8,776,796 |
|
|
|
|
20b |
RP11-684P7 |
BES |
chr20:5,395,692-5,580,006 |
|
|
|
|
20b |
CH250-20N7 |
|
chr20:3,593,189-3,753,758 |
|
macaque clone
|
|
|
20b |
RP11-867B19 |
BES |
chr20:2,754,801-2,928,115 |
20p13 |
|
|
|
- |
|
|
|
|
|
|
|
- |
RP11-794O20 |
BES |
chr20:24,399,808-24,587,032 |
|
Duplications
|
|
|
- |
RP11-475N20 |
BES |
chr20:24,635,369-24,808,231 |
|
|
|
- |
RP11-90H19 |
BES |
chr20:25,523,327-25,670,069 |
|
|
|
- |
|
|
|
|
|
|
|
- |
RP11-99E14 |
|
chr20:291,443-453,894 |
|
|
|
|
|
CH250-124B17 |
|
chr20:92,139-272,793 |
|
macaque clone
|
|
|
|
|
|
|
|
GGO PTR HSA breakpoint
|
|
|
20d |
RP11-659L8 |
BES |
chr20:29,831,888-30,017,666 |
20q11.21 |
|
|
|
20d |
RP11-597C24 |
BES |
chr20:30,207,948-30,385,641 |
|
|
|
|
20d |
RP11-308J23 |
BES |
chr20:32,216,912-32,396,349 |
|
|
|
|
20d |
CH250-265J5 |
|
chr20:33,939,476-34,113,827 |
|
macaque clone
|
|
|
20d |
RP11-663D7 |
BES |
chr20:34,099,025-34,292,351 |
20q11.22 |
|
|
|
 |
neocentromere not shared with Papionini |
|
|
20c |
RP11-643H5 |
|
chr20:11,431,493-11,596,313 |
20p12.2 |
|
|
|
20c |
CH250-139E11 |
|
chr20:12,584,448-12,777,937 |
|
macaque clone
|
|
|
20c |
RP11-248G23 |
BES |
chr20:15,095,739-15,228,679 |
|
|
|
|
20c |
RP11-14I7 |
BES |
chr20:17,730,109-17,913,893 |
|
|
|
|
20c |
RP11-237O17 |
BES |
chr20:21,287,083-21,431,542 |
|
|
|
|
20c |
RP11-627C15 |
BES |
chr20:24,208,343-24,399,802 |
20p11.21 |
|
|
|
- |
RP11-794O20 |
BES |
chr20:24,399,808-24,587,032 |
|
Duplications
|
|
|
- |
RP11-475N20 |
BES |
chr20:24,635,369-24,808,231 |
|
|
|
- |
RP11-90H19 |
BES |
chr20:25,523,327-25,670,069 |
|
|
|
- |
|
|
|
|
|
|
|
20a |
RP11-867B19 |
BES |
chr20:2,754,801-2,928,115 |
20p13 |
|
|
|
20a |
CH250-212J18 |
|
chr20:2,567,309-2,738,373 |
|
macaque clone
|
|
|
20a |
RP11-262I12 |
BES |
chr20:1,504,631-1,681,464 |
|
|
|
|
20a |
RP11-43K22 |
BES |
chr20:1,197,645-1,370,481 |
|
duplicated on 2q
|
|
|
20a |
CH250-141G14 |
|
chr20:498,598-662,151 |
|
macaque clone
|
|
|
20a |
RP11-99E14 |
BES |
chr20:291,443-453,894 |
20p13 |
|
|
|
- |
|
|
|
|
|
|
|
20e |
RP11-663D7 |
BES |
chr20:34,099,025-34,292,351 |
20q11.22 |
|
|
|
20e |
CH250-147C22 |
|
chr20:34,194,864-34,360,907 |
|
macaque clone
|
|
|
20e |
RP11-381P2 |
BES |
chr20:34,292,423-34,452,273 |
|
duplicated on 2q
|
|
|
20e |
RP11-115H6 |
BES |
chr20:34,673,620-34,872,526 |
|
|
|
|
20e |
CH250-160J22 |
|
chr20:35,418,191-35,593,431 |
|
macaque clone
|
|
|
20e |
RP11-1152L20 |
BES |
chr20:35,651,140-35,776,134 |
20q11.23 |
|
|
|
- |
|
|
|
|
|
|
|
21a |
RP11-468N22 |
BES |
chr21:35,738,487-35,926,512 |
21q22.12 |
|
|
|
21a |
CH250-204O04 |
|
chr21:35,375,423-35,568,778 |
|
macaque clone
|
|
|
21a |
RP11-124K12 |
BES |
chr21:24,834,561-24,978,580 |
|
|
|
|
21a |
RP11-300M18 |
BES |
chr21:18,875,076-19,039,028 |
|
|
|
|
21a |
RP11-138O15 |
BES |
chr21:15,627,586-15,772,076 |
21q11.2 |
|
|
|
- |
|
|
|
|
|
|
|
21b |
RP11-468N22 |
BES |
chr21:35,738,487-35,926,512 |
21q22.12 |
|
|
|
21b |
CH250-53F5 |
|
chr21:35,734,349-35,899,885 |
|
macaque clone
|
|
|
21b |
RP11-476D17 |
BES |
chr21:39,682,968-39,863,671 |
|
|
|
|
21b |
RP11-162F19 |
BES |
chr21:46,532,032-46,691,772 |
21q22.3 |
|
|