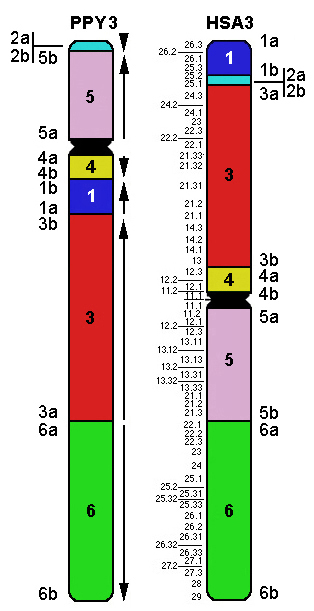
|
PPY 3
|
|||||||
|
|
BAC | Acc. N. | map | UCSC March 2006 |
comments
|
||
 |
2a
|
RP11-732C9 | BES | 3p25.2 | chr3:12,441,757-12,649,037 |
splits
|
|
 |
2 | RP11-767C1 | BES | 3p25.2 | chr3:12,728,844-12,897,825 | ||
 |
2 | RP11-588P9 | AC018836 | 3p25.2 | chr3:12,791,778-12,976,492 | ||
 |
2 | RP11-536I6 | AC093496 | 3p25.2 | chr3:14,236,276-14,420,312 | ||
 |
2b
|
RP11-616M11 | AC090954 | 3p25.1 | chr3:15,045,785-15,213,797 |
overlapp to 3a
|
|
|
|
|
PPAE-267I3 | BES | chr3:15,054,243/chr3:131,094,127 | FISH on HSA and PPY | ||
| PPAE-36H14 | BES | chr3:15,071,664/chr3:131,040,578 | FISH on HSA and PPY | ||||
 |
5b | RP11-924M2 | BES | 3q22.1 | chr3:131,126,893-131,354,295 | signals also on other chrs | |
 |
5
|
RP11-305I9 | AC092981 | 3q13.33 | chr3:120,465,181-120,625,176 | ||
 |
5a
|
RP11-454H13 | AC084198 | 3q12.3 | chr3:102,798,688-102,993,642 | ||
 |
PPY and HSA centromere | ||||||
 |
4a
|
RP11-601J5 | BES | 3p12.3 | chr3:75,877,002-76,036,449 | 0 kb from 3b | |
 |
4
|
RP11-536K4 | AC016942 | 3p12.3 | chr3:76,682,896-76,834,909 | ||
 |
4b
|
RP11-655A17 | AC107204 | 3p11.1 | chr3:87,099,705-87,270,490 | ||
|
|
|
PPAE-59o18 | BES | chr3:12,5/90.29 | |||
 |
1b
|
RP11-732C9 | BES | 3p25.2 | chr3:12,441,757-12,649,037 | splits | |
 |
1
|
RP11-183N22 | AL512885 (BLAT) | 3p26.1 | chr3:4,346,716-4,508,442 | ||
 |
1a
|
RP11-1120H22 | AC018810 | 3p26.3 | chr3:204,468-352,218 | ||
|
|
|
PPAE-143A06 | BES | chr3-:253,541/dup | FISH on HSA and PPY | ||
| PPAE-520C10 | BES | chr3-:209,308/dup | FISH on HSA and PPY | ||||
 |
3b
|
RP11-634L22 | BES | 3p12.3 | chr3:75,452,260-75,628,601 | 0 kb from 4a | |
 |
3
|
RP11-380J21 | AC092419 | 3p14.1 | chr3:64,053,486-64,212,751 | ||
 |
3
|
RP11-395P16 | AC130472 | 3p21.31 | chr3:47,584,747-47,778,906 | ||
 |
3
|
RP11-491D6 | AC006583 | 3p22.3 | chr3:37,034,150-37,136,520 | ||
 |
3
|
RP11-109D5 | AC074287 | 3p24.2 | chr3:25,497,571-25,696,055 | ||
 |
3a
|
RP11-421B21 | AC090949 | 3p25.1 | chr3:15,147,209-15,324,532 | overlaps to 2b | |
 |
6a
|
RP11-687B8 | AC112646 | 3q22.1 | chr3:131,420,953-131,459,232 | signals also on other chrs | |
 |
6
|
RP11-21N8 | AC023438 | 3q22.1 | chr3:131,810,596-131,961,213 | ||
 |
6
|
RP11-505J9 | AC024897 | 3q24 | chr3:149,845,113-150,049,906 | ||
 |
6
|
RP11-498P15 | AC112906 | 3q26.1 | chr3:163,530,323-163,648,093 | ||
 |
6
|
RP11-526M23 | AC048332 | 3q26.1 | chr3:166,898,263-167,089,798 | ||
 |
6
|
RP11-114M1 | AC026355 | 3q26.32 | chr3:178,755,557-178,912,996 | ||
 |
6
|
RP11-35G16 | AC016926 | 3q27.3 | chr3:187,694,918-187,861,239 | ||
 |
6
|
RP11-313F11 | AC016953 | 3q29 | chr3:196,907,957-197,370,697 | ||
 |
6b
|
HSA qtel | chr3:199,340,137 | ||||
HSA << great apes ancestor >> PPY
(Note that evolutionary synteny blocks are numbered according to their position in the ancestor,
while in the data above they are numbered according to their position in humans (see the Table below).
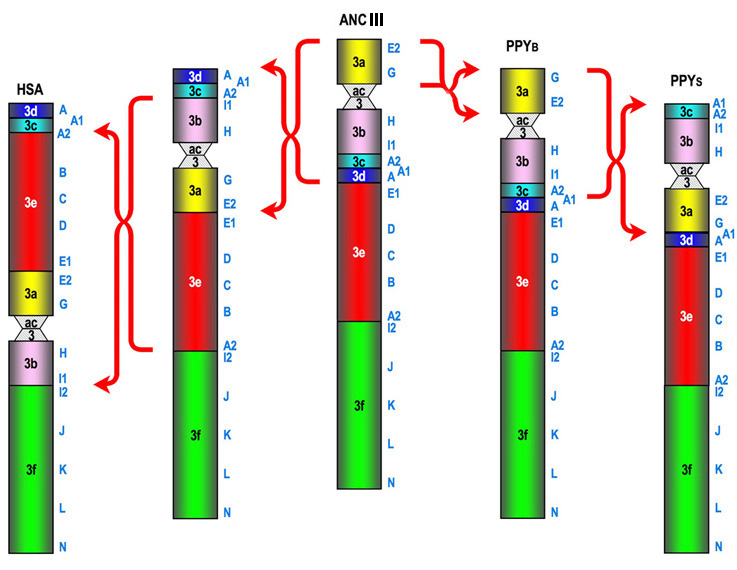
| code | BAC | Acc. N. | map | UCSC Mar. 2006 | comments | |
|
3d
|
A
|
RP11-183N22 | AL512885 | 3p26.1 | chr3:4346716-4508442 | |
|
3d/3c
|
A1
|
RP11-732C9 | BES | 3p25.2 | chr3:12,441,757-12,649,037 | split |
|
3c
|
A2
|
RP11-588P9 | AC018836 | 3p25.2 | chr3:12,791,778-12,976,492 | |
|
3c
|
B
|
RP11-109D5 | AC074287 | 3p24.2 | chr3:25,497,571-25,696,055 | |
|
3c
|
C
|
RP11-491D6 | AC006583 | 3p22.3 | chr3:37,034,150-37,136,520 | |
|
3c
|
D
|
RP11-395P16 | AC130472 | 3p21.31 | chr3:47,584,747-47,778,906 | |
|
3c
|
E1
|
RP11-634L22 | BES | 3p12.3 | chr3:75,452,260-75,628,601 | |
|
3a
|
E2
|
RP11-601J5 | BES | 3p12.3 | chr3:75,877,002-76,036,449 | |
|
3a
|
G
|
RP11-655A17 | AC107204 | 3p11.1 | chr3:87,099,705-87,270,490 | |
|
3b
|
H
|
RP11-454H13 | AC084198 | 3q12.3 | chr3:102,798,688-102,993,642 | |
|
3b
|
I1
|
RP11-924M2 | BES | 3q22.1 | chr3:131,126,893-131,354,295 | |
|
3f
|
I2
|
RP11-687B8 | AC112646 | 3q22.1 | chr3:131,420,953-131,459,232 | |
|
3f
|
J
|
RP11-21N8 | AC023438 | 3q22.1 | chr3:131,810,596-131,961,213 | |
|
3f
|
K
|
RP11-505J9 | AC024897 | 3q24 | chr3:149,845,113-150,049,906 | |
|
3f
|
L
|
RP11-526M23 | AC048332 | 3q26.1 | chr3:166,898,263-167,089,798 | |
|
3f
|
N
|
RP11-313F11 | AC016953 | 3q29 | chr3:196,907,957-197,370,697 |