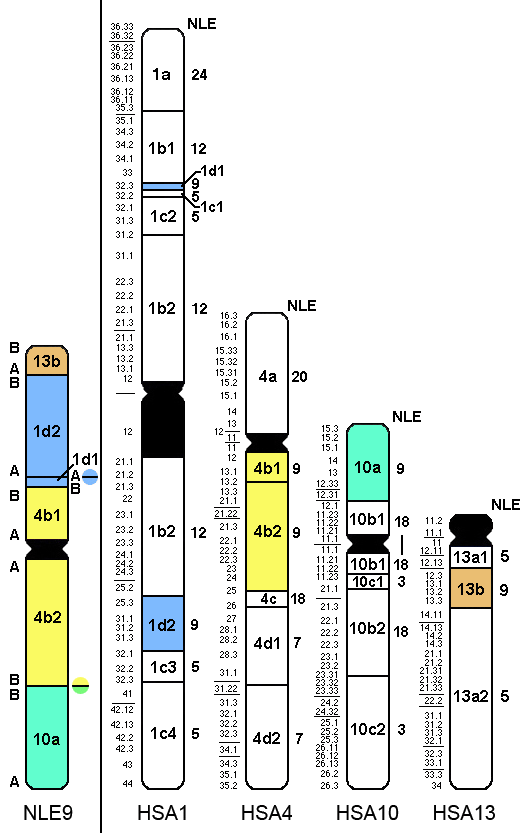
 |
|
|
segm.
|
clone |
Acc.N. |
UCSC May 2004
|
map |
note |
 |
B
|
13b
|
RP11-597B2 |
BES |
chr13:38,988,139-39,162,449 |
13q13.3 |
69kb overlap |
 |
|
13b
|
RP11-58M19 |
BES |
chr13:28,785,075-28,945,002 |
|
|
 |
A |
13b
|
RP11-780D7 |
BES |
chr13:26,335,941-26,501,574 |
13q12.13 |
splits |
|
|
-
|
CH271-134B7 |
BES |
chr1:200,305,408/chr13:26,359,161 |
|
splits |
 |
B |
1d2
|
RP11-397P13 |
BES |
chr1:200,391,715-200,571,022 |
1q32.1 |
splits |
 |
|
1d2 |
RP11-813P3 |
BES |
chr1:200,111,287-200,347,082 |
|
|
 |
|
1d2
|
RP11-173E24 |
BES |
chr1:191,987,584-192,156,745 |
|
|
 |
A
|
1d2
|
RP11-453M18 |
AL157408 |
chr1:177,774,617-177,954,344 |
1q25.3 |
splits |
|
|
-
|
CH271-208N2 |
|
chr1:177,996,656/chr1:52,346,210 |
|
|
 |
A
|
1d1
|
RP11-978G17 |
BES |
chr1:52,122,191-52,296,879 |
1p32.3 |
splits |
 |
|
1d1 |
RP11-593L12 |
BES |
chr1:52,563,756-52,734,413 |
|
|
 |
B |
1d1
|
RP11-844G13 |
BES |
chr1:53,401,173-53,597,036 |
1p32.3 |
9kb overlap |
|
|
-
|
|
|
|
|
|
|
|
|
|
|
duplications |
 |
B |
4b1
|
RP11-245E9 |
BES |
chr4:69,547,000-69,700,016 |
4q13.2 |
129kb |
 |
|
4b1 |
RP11-139N1 |
BES |
chr4:61,407,224-61,546,488 |
|
|
 |
A
|
4b1
|
RP11-365H22 |
AC027271 |
chr4:52,501,046-52,679,030 |
4q11 |
cen |
|
 |
|
centromere |
|
|
|
|
|
|
|
|
|
duplications |
 |
A |
4b2 |
RP11-915G24 |
BES |
chr4:69,789,054-69,958,759 |
4q13.2 |
129kb |
 |
|
4b2
|
RP11-203P17 |
BES |
chr4:70,246,613-70,431,315 |
|
|
 |
|
4b2
|
RP11-499E18 |
AC098487 |
chr4:103,572,839-103,736,754 |
|
|
 |
B
|
4b2
|
RP11-1091M15 |
BES |
chr4:110,517,162-110,721,162 |
4q25 |
splits |
|
|
-
|
CH271-138C4 |
|
chr4:110,472,197/chr10:23,987,314 |
|
splits |
 |
B |
10a
|
RP11-344N19 |
BES |
chr10:23,942,660-24,115,999 |
10p12.2 |
splits |
 |
|
10a
|
RP11-368M8 |
BES |
chr10:22,833,626-23,007,256 |
|
|
 |
A |
10a
|
RP11-363N22 |
AL359878 |
chr10:854,871-1,039,159 |
10p15.3 |
854kb/tel |
|

